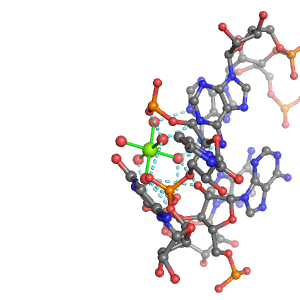
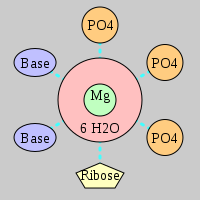
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3i1t |
A3028 |
3Pout·Rout·2Bout |
|
|
|
A3594, A3595, A3596, A3597, A3598, A3599 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, A:A821 |
|
3oas |
A3027 |
3Pout·Rout·2Bout |
|
|
|
A3593, A3594, A3595, A3596, A3597, A3598 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, U:A568 |
|
3ofd |
A3027 |
3Pout·Rout·2Bout |
|
|
|
A3594, A3595, A3596, A3597, A3598, A3599 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, U:A568 |
|
3ofq |
A3028 |
3Pout·Rout·2Bout |
|
|
|
A3594, A3595, A3596, A3597, A3598, A3599 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, U:A568 |
|
3ofr |
A3032 |
3Pout·Rout·2Bout |
|
|
|
A3610, A3611, A3612, A3613, A3614, A3615 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, A:A821 |
|
3oxd |
A101 |
3Pout·Rout·2Bout |
|
|
|
A138, A139, A140, A141, A142, A143 |
|
C:A35, A:A65, A:A34 |
A:A81 |
G:A36, A:A37 |
|
3oxd |
B106 |
3Pout·Rout·2Bout |
|
|
|
A144, A145, B137, B138, B139, B140 |
|
C:A66, C:B66, A:B33 |
A:B65 |
A:B33, A:A65 |
|
3r8s |
A3033 |
3Pout·Rout·2Bout |
|
|
|
A3680, A3681, A3682, A3683, A3684, A3685 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, A:A821 |
|
3r8t |
A2950 |
3Pout·Rout·2Bout |
|
|
|
A3286, A3287, A3288, A3289, A3290 |
|
G:A1455, A:A1453, G:A1456 |
C:A1454 |
G:A1456, U:A1457 |
|
3r8t |
A3032 |
3Pout·Rout·2Bout |
|
|
|
A3649, A3650, A3651, A3652, A3653, A3654 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, A:A821 |
|
3v23 |
A3606 |
3Pout·Rout·2Bout |
|
|
|
A5030, A5031, A5032, A5033, A5034, A5035 |
|
G:A570, A:A973, U:A569 |
G:A972 |
A:A820, A:A821 |
|
3v27 |
A3629 |
3Pout·Rout·2Bout |
|
|
|
A5230, A5231, A5232, A5233, A5234, A5235 |
|
G:A570, A:A973, U:A569 |
G:A972 |
A:A820, A:A821 |
|
3v2f |
A3192 |
3Pout·Rout·2Bout |
|
|
|
A3894, A3895, A3896, A3897 |
|
A:A1937, U:A1956, G:A1945 |
A:A1936 |
G:A1945, U:A1946 |
|
4bpe |
A5063 |
3Pout·Rout·2Bout |
|
|
|
A2372, A2373, A2374, A2375, A2434, A2438 |
|
G:A1173, G:A1172, G:A1170 |
C:A1574 |
A:A1168, C:A1574 |
|
4bpn |
A5063 |
3Pout·Rout·2Bout |
|
|
|
A6507, A6508, A6509, A6510, A6511, A6512 |
|
G:A1173, G:A1170, G:A1172 |
C:A1574 |
A:A1168, C:A1574 |
|
4bpo |
A5063 |
3Pout·Rout·2Bout |
|
|
|
A2220, A2221, A2222, A2223, A2294, A2298 |
|
G:A1173, G:A1172, G:A1170 |
C:A1574 |
A:A1168, C:A1574 |
|
4gar |
A3127 |
3Pout·Rout·2Bout |
|
|
|
A3775, A3776, A3777, A3778, A3779, A3780 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A821, A:A820 |
|
4gau |
A3126 |
3Pout·Rout·2Bout |
|
|
|
A3779, A3780, A3781, A3782, A3783, A3784 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, A:A821 |
|
4ji8 |
A1959 |
3Pout·Rout·2Bout |
|
|
|
A3258, A3259, A3260, A3261, A3262, A3263 |
|
C:A857, G:A869, C:A868 |
G:A858 |
G:A858, G:A869 |
|
4peb |
A3129 |
3Pout·Rout·2Bout |
|
|
|
A3778, A3779, A3780, A3781, A3782, A3783 |
|
G:A570, A:A973, U:A569 |
A:A972 |
A:A820, A:A821 |
|
4qcr |
A3498 |
3Pout·Rout·2Bout |
|
|
|
A4618, A4619, A4620, A4621, A4622, A4623 |
|
G:A593, A:A1018, U:A592 |
G:A1017 |
A:A867, A:A868 |
|
4qct |
A3389 |
3Pout·Rout·2Bout |
|
|
|
A4387, A4388, A4389, A4390, A4391, A4392 |
|
G:A570, U:A569, A:A973 |
G:A972 |
A:A820, A:A821 |
|
4qcx |
A3389 |
3Pout·Rout·2Bout |
|
|
|
A4320, A4321, A4322, A4323, A4324, A4325 |
|
G:A570, A:A973, U:A569 |
G:A972 |
A:A820, A:A821 |
|
4qcz |
A3479 |
3Pout·Rout·2Bout |
|
|
|
A4562, A4563, A4564, A4565, A4566 |
|
U:A592, G:A593, A:A1018 |
G:A1017 |
A:A867, A:A868 |
|
4qd1 |
A3402 |
3Pout·Rout·2Bout |
|
|
|
A4310, A4311, A4312, A4313, A4314, A4315 |
|
G:A570, U:A569, A:A973 |
G:A972 |
A:A820, A:A821 |
|