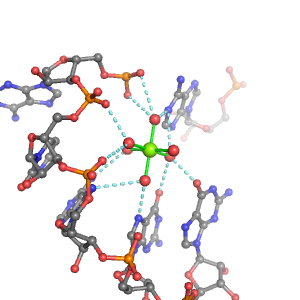
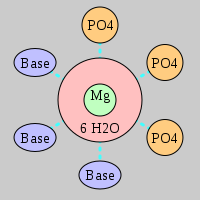
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1dul |
B801 |
3Pout·3Bout |
|
|
|
B810, B820, B826, B832, B881, B882 |
|
C:B163, G:B144, G:B145 |
|
U:B146, G:B144, G:B145 |
|
1hq1 |
B1801 |
3Pout·3Bout |
|
|
|
B9011, B9015, B9025, B9028, B9038, B9180 |
|
C:B163, G:B144, G:B145 |
|
U:B146, G:B144, G:B145 |
|
1vs6 |
B2983 |
3Pout·3Bout |
|
|
|
B3388, B3389, B3390, B3391, B3392, B3393 |
|
A:B2451, C:B2452, A:B2453 |
|
G:B2454, A:B2497, G:B2455 |
|
1vs8 |
B2917 |
3Pout·3Bout |
|
|
|
B3075, B3076, B3077, B3078, B3079, B3080 |
|
A:B2033, G:B2027, U:B2026 |
|
G:B2027, U:B2028, G:B2029 |
|
1vs8 |
B2958 |
3Pout·3Bout |
|
|
|
B3273, B3274, B3275, B3276, B3277, B3278 |
|
A:B582, G:B583, A:B1253 |
|
C:B584, A:B582, G:B583 |
|
1vt2 |
A2906 |
3Pout·3Bout |
|
|
|
A3044, A3045, A3046, A3047, A3048, A3049 |
|
A:A28, U:A29, A:A447 |
|
G:A473, U:A29, G:A30 |
|
2awb |
B2917 |
3Pout·3Bout |
|
|
|
B3076, B3077, B3078, B3079, B3080, B3081 |
|
A:B2033, G:B2027, U:B2026 |
|
G:B2027, U:B2028, G:B2029 |
|
2i2t |
B2957 |
3Pout·3Bout |
|
|
|
B3274, B3275, B3276, B3277, B3278, B3279 |
|
A:B616, G:B617, A:B608 |
|
G:B617, G:B619, G:B618 |
|
2i2v |
B2939 |
3Pout·3Bout |
|
|
|
B3194, B3195, B3196, B3197, B3198, B3199 |
|
A:B447, A:B28, U:B29 |
|
U:B29, G:B30, G:B474 |
|
2i2v |
B2927 |
3Pout·3Bout |
|
|
|
B3134, B3135, B3136, B3137, B3138, B3139 |
|
G:B617, A:B608, A:B616 |
|
G:B618, G:B617, G:B619 |
|
2qou |
A1584 |
3Pout·3Bout |
|
|
|
A2098, A2099, A2100, A2101, A2102 |
|
A:A1110, A:A1111, C:A1109 |
|
U:A1189, A:A1111, A:A1188 |
|
2qov |
B2923 |
3Pout·3Bout |
|
|
|
B3103, B3104, B3105, B3106, B3107, B3108 |
|
U:B29, A:B28, A:B447 |
|
G:B473, G:B30, G:B474 |
|
3df2 |
B3110 |
3Pout·3Bout |
|
|
|
B4111, B4112, B4113, B4114, B4115, B4116 |
|
U:B29, A:B28, A:B447 |
|
G:B473, G:B474, G:B30 |
|
3df4 |
B3207 |
3Pout·3Bout |
|
|
|
B4208, B4209, B4210, B4211, B4212, B4213 |
|
A:B447, A:B28, U:B29 |
|
G:B30, G:B473, G:B474 |
|
3i1o |
A2 |
3Pout·3Bout |
|
|
|
A1577, A1578, A1579, A1580, A1581, A1582 |
|
A:A329, A:A315, G:A319 |
|
G:A319, A:A320, G:A318 |
|
3i1r |
A2974 |
3Pout·3Bout |
|
|
|
A3369, A3370, A3371, A3372, A3373, A3374 |
|
A:A2453, A:A2451, C:A2452 |
|
A:A2497, G:A2455, A:A2453 |
|
3i1t |
A2905 |
3Pout·3Bout |
|
|
|
A3044, A3045, A3046, A3047, A3048, A3049 |
|
U:A29, A:A28, A:A447 |
|
G:A473, G:A474, G:A30 |
|
3i20 |
A3036 |
3Pout·3Bout |
|
|
|
A3638, A3639, A3640, 4779, 4782, 4784 |
|
A:A2740, A:A2741, G:A2742 |
|
G:A2742, G:A2763, U:A2743 |
|
3oar |
A1535 |
3Pout·3Bout |
|
|
|
A1577, A1578, A1579, A1580, A1581, A1582 |
|
A:A315, A:A329, G:A319 |
|
G:A319, A:A320, G:A318 |
|
3ofb |
A1535 |
3Pout·3Bout |
|
|
|
A1578, A1579, A1580, A1581, A1582, A1583 |
|
A:A315, A:A329, G:A319 |
|
G:A319, A:A320, G:A318 |
|
3ofy |
A1535 |
3Pout·3Bout |
|
|
|
A1578, A1579, A1580, A1581, A1582, A1583 |
|
A:A315, A:A329, G:A319 |
|
G:A319, A:A320, G:A318 |
|
3orb |
A2906 |
3Pout·3Bout |
|
|
|
A3046, A3047, A3048, A3049, A3050, A3051 |
|
A:A28, U:A29, A:A447 |
|
G:A473, G:A474, U:A29 |
|
3ucz |
R1 |
3Pout·3Bout |
|
|
|
R671, R672, R673, R674, R675, R676 |
|
A:R23, G:R41, G:R40 |
|
G:R42, A:R43, G:R41 |
|
3v23 |
A3605 |
3Pout·3Bout |
|
|
|
A5024, A5025, A5026, A5027, A5028, A5029 |
|
A:A2451, C:A2452, A:A2453 |
|
A:A2453, A:A2497, G:A2454 |
|
4bpe |
A5069 |
3Pout·3Bout |
|
|
|
A2368, A2369, A2370, A2371, A2376, A2377 |
|
G:A1148, C:A1428, C:A1149 |
|
A:A1174, U:A1159, G:A1172 |
|