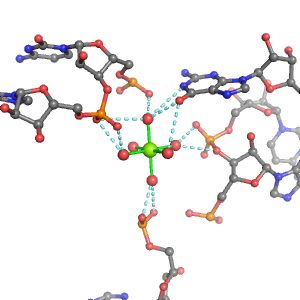
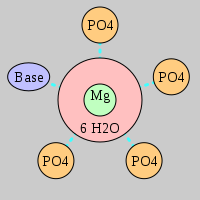
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2qp1 |
B2910 |
4Pout·Bout |
|
|
|
B3548, B3549, B3550, B3551, B3552, B3553 |
|
A:B1987, C:B1986, C:B2551, A:B1669 |
|
G:B1954 |
|
2z4l |
B2936 |
4Pout·Bout |
|
|
|
B3160, B3161, B3162, B3163, B3164, B3165 |
|
U:B2613, C:B2050, A:B2051, G:B2578 |
|
A:B2614 |
|
2z4l |
B2971 |
4Pout·Bout |
|
|
|
B3324, B3325, B3326, B3327, B3328, B3329 |
|
A:B1669, C:B1986, A:B1987, C:B2551 |
|
G:B1954 |
|
2z4n |
B2911 |
4Pout·Bout |
|
|
|
B3043, B3044, B3045, B3046, B3047, B3048 |
|
C:B2551, A:B1987, A:B1669, C:B1986 |
|
G:B1954 |
|
3df2 |
B3181 |
4Pout·Bout |
|
|
|
B4182, B4183, B4184, B4185, B4186, B4187 |
|
U:B2613, C:B2050, A:B2051, G:B2578 |
|
A:B2614 |
|
3df2 |
B3382 |
4Pout·Bout |
|
|
|
B4383, B4384, B4385, B4386, B4387, B4388 |
|
A:B1669, C:B1986, C:B2551, A:B1987 |
|
G:B1954 |
|
3df4 |
B3033 |
4Pout·Bout |
|
|
|
B4034, B4035, B4036, B4037, B4038, B4039 |
|
C:B1986, C:B2551, A:B1669, A:B1987 |
|
G:B1954 |
|
3df4 |
B3026 |
4Pout·Bout |
|
|
|
B4027, B4028, B4029, B4030, B4031, B4032 |
|
A:B1937, A:B1936, C:B1958, G:B1959 |
|
G:B1959 |
|
3i1n |
A2947 |
4Pout·Bout |
|
|
|
A3243, A3244, A3245, A3246, A3247, A3248 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|
3i1n |
A2954 |
4Pout·Bout |
|
|
|
A3275, A3276, A3277, A3278, A3279, A3280 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3i1p |
A2956 |
4Pout·Bout |
|
|
|
A3279, A3280, A3281, A3282, A3283, A3284 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3i1p |
A2949 |
4Pout·Bout |
|
|
|
A3247, A3248, A3249, A3250, A3251, A3252 |
|
C:A1351, C:A1376, U:A1352, G:A1377 |
|
G:A1377 |
|
3i1r |
A2954 |
4Pout·Bout |
|
|
|
A3272, A3273, A3274, A3275, A3276, A3277 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3i1r |
A2947 |
4Pout·Bout |
|
|
|
A3240, A3241, A3242, A3243, A3244, A3245 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|
3i1t |
A2956 |
4Pout·Bout |
|
|
|
A3282, A3283, A3284, A3285, A3286, A3287 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3i1t |
A2949 |
4Pout·Bout |
|
|
|
A3250, A3251, A3252, A3253, A3254, A3255 |
|
C:A1351, C:A1376, U:A1352, G:A1377 |
|
G:A1377 |
|
3i20 |
A2947 |
4Pout·Bout |
|
|
|
A3241, A3242, A3243, A3244, A3245, A3246 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|
3i20 |
A2954 |
4Pout·Bout |
|
|
|
A3273, A3274, A3275, A3276, A3277, A3278 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3i22 |
A2947 |
4Pout·Bout |
|
|
|
A3244, A3245, A3246, A3247, A3248 |
|
C:A1314, G:A1332, G:A1334, U:A1313 |
|
C:A1335 |
|
3mut |
R671 |
4Pout·Bout |
|
|
|
R715, R716, R717, R718, R719, R720 |
|
U:R53, U:R81, C:R52, C:R44 |
|
A:R82 |
|
3oas |
A2948 |
4Pout·Bout |
|
|
|
A3248, A3249, A3250, A3251, A3252, A3253 |
|
C:A1351, C:A1376, U:A1352, G:A1377 |
|
G:A1377 |
|
3oas |
A2955 |
4Pout·Bout |
|
|
|
A3280, A3281, A3282, A3283, A3284, A3285 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3oat |
A2953 |
4Pout·Bout |
|
|
|
A3264, A3265, A3266, A3267, A3268, A3269 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3ofc |
A2952 |
4Pout·Bout |
|
|
|
A3266, A3267, A3268, A3269, A3270, A3271 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3ofd |
A2955 |
4Pout·Bout |
|
|
|
A3278, A3279, A3280, A3281, A3282, A3283 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|