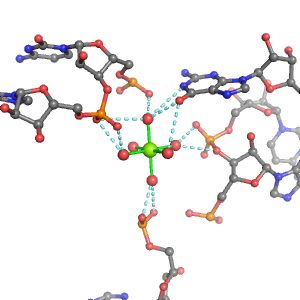
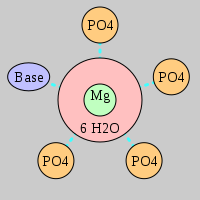
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3ofq |
A2949 |
4Pout·Bout |
|
|
|
A3249, A3250, A3251, A3252, A3253, A3254 |
|
C:A1351, C:A1376, U:A1352, G:A1377 |
|
G:A1377 |
|
3ofq |
A2956 |
4Pout·Bout |
|
|
|
A3281, A3282, A3283, A3284, A3285, A3286 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3ofr |
A2954 |
4Pout·Bout |
|
|
|
A3269, A3270, A3271, A3272, A3273, A3274 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3ofz |
A2945 |
4Pout·Bout |
|
|
|
A3230, A3231, A3232, A3233, A3234, A3235 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|
3ofz |
A2952 |
4Pout·Bout |
|
|
|
A3262, A3263, A3264, A3265, A3266, A3267 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3og0 |
A2947 |
4Pout·Bout |
|
|
|
A3246, A3247, A3248, A3249, A3250, A3251 |
|
C:A1351, C:A1376, U:A1352, G:A1377 |
|
G:A1377 |
|
3og0 |
A2954 |
4Pout·Bout |
|
|
|
A3278, A3279, A3280, A3281, A3282, A3283 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3orb |
A2953 |
4Pout·Bout |
|
|
|
A3269, A3270, A3271, A3272, A3273, A3274 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
3orb |
A2946 |
4Pout·Bout |
|
|
|
A3237, A3238, A3239, A3240, A3241, A3242 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|
3r8s |
A2954 |
4Pout·Bout |
|
|
|
A3328, A3329, A3330, A3331, A3332, A3333 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3r8t |
A2946 |
4Pout·Bout |
|
|
|
A3269, A3270, A3271, A3272, A3273, A3274 |
|
U:A1352, C:A1376, G:A1377, C:A1351 |
|
G:A1377 |
|
3r8t |
A2953 |
4Pout·Bout |
|
|
|
A3301, A3302, A3303, A3304, A3305, A3306 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
3ucz |
R670 |
4Pout·Bout |
|
|
|
R687, R688, R689, R690, R691, R692 |
|
U:R53, C:R52, U:R81, C:R44 |
|
A:R82 |
|
3v23 |
A3579 |
4Pout·Bout |
|
|
|
A4889, A4890, A4891, A4892, A4893 |
|
A:A1986, C:A2551, A:A1669, G:A1987 |
|
G:A1954 |
|
3v28 |
A1749 |
4Pout·Bout |
|
|
|
A2007, A2008, A2009, A2010, A2011 |
|
A:A16, U:A13, U:A14, G:A15 |
|
U:A14 |
|
3v29 |
A3571 |
4Pout·Bout |
|
|
|
A4655, A4656, A4657, A4658, A4659, A4660 |
|
G:A1987, A:A1669, A:A1986, C:A2551 |
|
G:A1954 |
|
3v2d |
A3223 |
4Pout·Bout |
|
|
|
A4312, A4313, A4314, A4315 |
|
C:A2745, U:A2746, U:A2754, C:A2755 |
|
G:A2747 |
|
3v2f |
A3599 |
4Pout·Bout |
|
|
|
A5065, A5066, A5067, A5068, A5069, A5070 |
|
G:A1987, A:A1669, A:A1986, C:A2551 |
|
G:A1954 |
|
4bpe |
A5002 |
4Pout·Bout |
|
|
|
A2164, A2165, A2188, A2241, A2343, A2344 |
|
A:A919, G:A920, A:A1005, A:A1743 |
|
A:A1003 |
|
4bpn |
A5012 |
4Pout·Bout |
|
|
|
A6317, A6319, A6326, A6333, A6340, A6347 |
|
A:A969, G:A989, C:A988, C:A968 |
|
G:A989 |
|
4bpo |
A5012 |
4Pout·Bout |
|
|
|
A2178, A2179, A2180, A2186, A2187, A2341 |
|
C:A968, G:A989, A:A969, C:A988 |
|
G:A989 |
|
4dr6 |
A1840 |
4Pout·Bout |
|
|
|
A2505, A2506, A2507, A2508, A2509, A2510 |
|
A:A151, C:A150, G:A168, G:A167 |
|
G:A168 |
|
4dr6 |
A1608 |
4Pout·Bout |
|
|
|
A2019, A2020, A2021, A2022 |
|
U:A751, U:A652, G:A752, C:A651 |
|
G:A752 |
|
4dr7 |
A1818 |
4Pout·Bout |
|
|
|
A2474, A2475, A2476, A2477, A2478, A2479 |
|
C:A150, G:A168, A:A151, G:A167 |
|
G:A168 |
|
4duy |
A1793 |
4Pout·Bout |
|
|
|
A2143, A2144, A2145, A2146, A2147, A2148 |
|
A:A151, G:A167, G:A168, C:A150 |
|
G:A168 |
|