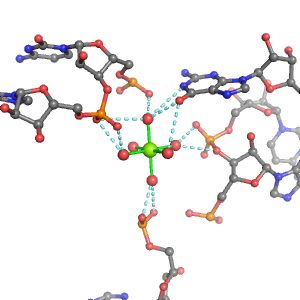
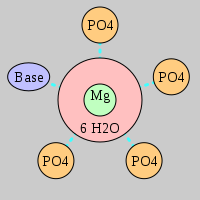
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4duz |
A1829 |
4Pout·Bout |
|
|
|
A2198, A2199, A2200, A2201, A2202, A2203 |
|
A:A509, A:A510, G:A506, C:A507 |
|
A:A510 |
|
4dv1 |
A1812 |
4Pout·Bout |
|
|
|
A2217, A2218, A2219, A2220, A2221 |
|
A:A509, A:A510, G:A506, C:A507 |
|
A:A510 |
|
4dv3 |
A1837 |
4Pout·Bout |
|
|
|
A2201, A2202, A2203, A2204, A2205, A2206 |
|
A:A509, A:A510, C:A507, G:A506 |
|
A:A510 |
|
4dv6 |
A1804 |
4Pout·Bout |
|
|
|
A2155, A2156, A2157, A2158, A2159, A2160 |
|
A:A151, G:A168, G:A167, C:A150 |
|
G:A168 |
|
4dv7 |
A1829 |
4Pout·Bout |
|
|
|
A2115, A2116, A2117, A2118, A2119, A2120 |
|
C:A150, A:A151, G:A168, G:A167 |
|
G:A168 |
|
4gar |
A3008 |
4Pout·Bout |
|
|
|
A3243, A3244, A3245, A3246 |
|
A:A471, A:A472, A:A449, A:A453 |
|
G:A450 |
|
4gar |
C301 |
4Pout·Bout |
|
|
|
A3747, A3748, A3749, C406, C407 |
|
G:A1828, U:A1827, A:A1789, C:A1788 |
|
G:A1828 |
|
4gar |
A3055 |
4Pout·Bout |
|
|
|
A3459, A3460, A3461, A3462, A3463, A3464 |
|
A:A1936, A:A1937, C:A1958, G:A1959 |
|
G:A1959 |
|
4gau |
A3048 |
4Pout·Bout |
|
|
|
A3427, A3428, A3429, A3430, A3431, A3432 |
|
A:A1669, C:A1986, C:A2551, A:A1987 |
|
G:A1954 |
|
4gau |
A3117 |
4Pout·Bout |
|
|
|
A3745, A3746, A3747, A3748 |
|
C:A192, U:A2243, C:A198, A:A199 |
|
A:A195 |
|
4ji1 |
A1797 |
4Pout·Bout |
|
|
|
A2473, A2474, A2475, A2476, A2477, A2478 |
|
C:A150, G:A168, G:A167, A:A151 |
|
G:A168 |
|
4ji1 |
A1730 |
4Pout·Bout |
|
|
|
A2070, A2071, A2072, A2073, A2074, A2075 |
|
U:A751, G:A752, U:A652, C:A651 |
|
G:A752 |
|
4ji8 |
A1951 |
4Pout·Bout |
|
|
|
A3211, A3212, A3213, A3214, A3215, A3216 |
|
C:A150, G:A167, G:A168, A:A151 |
|
G:A168 |
|
4nvw |
A3083 |
4Pout·Bout |
|
|
|
A4021, A4022, A4023, A4024 |
|
A:A768, U:A804, C:A805, A:A767 |
|
A:A768 |
|
4nvx |
A3553 |
4Pout·Bout |
|
|
|
A4700, A4701, A4702, A4703, A4704, A4705 |
|
A:A2051, A:A2614, C:A2050, G:A2578 |
|
A:A2051 |
|
4nvz |
A3449 |
4Pout·Bout |
|
|
|
A4471, A4472, A4473, A4474 |
|
A:A2008, C:A2563, A:A1716, G:A2009 |
|
G:A1976 |
|
4nw0 |
A3083 |
4Pout·Bout |
|
|
|
A4021, A4022, A4023, A4024 |
|
A:A768, U:A804, C:A805, A:A767 |
|
A:A768 |
|
4nw1 |
A3547 |
4Pout·Bout |
|
|
|
A4699, A4700, A4701, A4702, A4703, A4704 |
|
A:A2051, A:A2614, C:A2050, G:A2578 |
|
A:A2051 |
|
4peb |
A3050 |
4Pout·Bout |
|
|
|
A3430, A3431, A3432, A3433, A3434, A3435 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
4peb |
A3043 |
4Pout·Bout |
|
|
|
A3398, A3399, A3400, A3401, A3402, A3403 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|
4pec |
A3043 |
4Pout·Bout |
|
|
|
A3390, A3391, A3392, A3393, A3394, A3395 |
|
U:A1352, G:A1377, C:A1376, C:A1351 |
|
G:A1377 |
|
4pec |
A3050 |
4Pout·Bout |
|
|
|
A3422, A3423, A3424, A3425, A3426, A3427 |
|
A:A1669, C:A1986, A:A1987, C:A2551 |
|
G:A1954 |
|
4pec |
A3039 |
4Pout·Bout |
|
|
|
A3378, A3379, A3380, A3381 |
|
A:A2005, C:A2006, A:A1268, A:A1269 |
|
A:A1269 |
|
4qcz |
A3432 |
4Pout·Bout |
|
|
|
A4446, A4447, A4448, A4449 |
|
A:A2008, C:A2563, A:A1716, G:A2009 |
|
G:A1976 |
|
4tom |
A3043 |
4Pout·Bout |
|
|
|
A3395, A3396, A3397, A3398, A3399, A3400 |
|
C:A1351, U:A1352, G:A1377, C:A1376 |
|
G:A1377 |
|