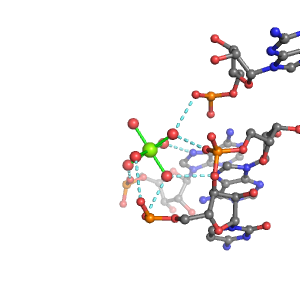
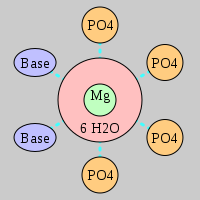
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
1vs6 |
B2935 |
4Pout·2Bout |
|
|
|
B3163, B3164, B3165, B3166, B3167, B3168 |
|
C:B2050, A:B2051, G:B2578, A:B2614 |
|
A:B2051, A:B2614 |
|
2qan |
A2189 |
4Pout·2Bout |
|
|
|
A2190, A2191, A2192, A2193, A2194, A2195 |
|
A:A768, U:A804, C:A805, A:A767 |
|
A:A768, G:A769 |
|
2qao |
B3026 |
4Pout·2Bout |
|
|
|
B3639, B3640, B3641, B3642, B3643, B3644 |
|
C:B1958, G:B1959, A:B1936, A:B1937 |
|
A:B1960, G:B1959 |
|
2qba |
B3181 |
4Pout·2Bout |
|
|
|
B3759, B3760, B3761, B3762, B3763, B3764 |
|
C:B2050, G:B2578, A:B2051, A:B2614 |
|
A:B2051, A:B2614 |
|
2qba |
B3157 |
4Pout·2Bout |
|
|
|
B3739, B3740, B3741, B3742, B3743, B3744 |
|
A:B1937, A:B1936, C:B1958, G:B1959 |
|
G:B1959, A:B1960 |
|
2qbc |
B3026 |
4Pout·2Bout |
|
|
|
B3640, B3641, B3642, B3643, B3644, B3645 |
|
A:B1937, C:B1958, A:B1936, G:B1959 |
|
A:B1960, G:B1959 |
|
2qbe |
B3181 |
4Pout·2Bout |
|
|
|
B3759, B3760, B3761, B3762, B3763, B3764 |
|
C:B2050, G:B2578, A:B2051, A:B2614 |
|
A:B2051, A:B2614 |
|
2qbf |
A1572 |
4Pout·2Bout |
|
|
|
A1757, A1758, A1759, A1760, A1761, A1762 |
|
A:A768, U:A804, C:A805, A:A767 |
|
G:A769, A:A768 |
|
2qbi |
B2931 |
4Pout·2Bout |
|
|
|
B3141, B3142, B3143, B3144, B3145, B3146 |
|
G:B1959, A:B1937, A:B1936, C:B1958 |
|
G:B1959, A:B1960 |
|
2qow |
A1560 |
4Pout·2Bout |
|
|
|
A1972, A1973, A1974, A1975, A1976, A1977 |
|
A:A768, U:A804, C:A805, A:A767 |
|
G:A769, A:A768 |
|
2qox |
B2909 |
4Pout·2Bout |
|
|
|
B3035, B3036, B3037, B3038, B3039, B3040 |
|
A:B1936, A:B1937, G:B1959, C:B1958 |
|
G:B1959, A:B1960 |
|
2qoz |
B2931 |
4Pout·2Bout |
|
|
|
B3636, B3637, B3638, B3639, B3640, B3641 |
|
G:B1959, A:B1937, A:B1936, C:B1958 |
|
G:B1959, A:B1960 |
|
2qp1 |
B2909 |
4Pout·2Bout |
|
|
|
B3542, B3543, B3544, B3545, B3546, B3547 |
|
A:B1937, G:B1959, A:B1936, C:B1958 |
|
G:B1959, A:B1960 |
|
2z4m |
A1573 |
4Pout·2Bout |
|
|
|
A1755, A1756, A1757, A1758, A1759, A1760 |
|
A:A768, U:A804, C:A805, A:A767 |
|
G:A769, A:A768 |
|
3df1 |
A2044 |
4Pout·2Bout |
|
|
|
A4045, A4046, A4047, A4048, A4049, A4050 |
|
A:A329, A:A315, U:A317, C:A316 |
|
G:A318, G:A319 |
|
3df3 |
A2189 |
4Pout·2Bout |
|
|
|
A4190, A4191, A4192, A4193, A4194, A4195 |
|
A:A768, C:A805, U:A804, A:A767 |
|
A:A768, G:A769 |
|
3r8s |
A2960 |
4Pout·2Bout |
|
|
|
A3358, A3359, A3360, A3361, A3362, A3363 |
|
C:A1958, G:A1959, A:A1936, A:A1937 |
|
G:A1959, A:A1960 |
|
3r8t |
A2959 |
4Pout·2Bout |
|
|
|
A3331, A3332, A3333, A3334, A3335, A3336 |
|
A:A1937, C:A1958, A:A1936, G:A1959 |
|
G:A1959, A:A1960 |
|
3v25 |
A3285 |
4Pout·2Bout |
|
|
|
A3693, A3694, A3695, A3696, A3697 |
|
A:A1634, G:A1627, G:A1628, U:A1300 |
|
G:A1628, U:A1629 |
|
3v28 |
A1752 |
4Pout·2Bout |
|
|
|
A2023, A2024, A2025, A2026, A2027, A2028 |
|
A:A780, G:A800, C:A779, G:A799 |
|
A:A780, G:A800 |
|
3v2d |
A3696 |
4Pout·2Bout |
|
|
|
A5676, A5677, A5678, A5679 |
|
G:A2578, A:A2051, U:A2613, A:A2614 |
|
A:A2051, A:A2614 |
|
3v2f |
A3601 |
4Pout·2Bout |
|
|
|
A5077, A5078, A5079, A5080, A5081, A5082 |
|
G:A2578, A:A2614, A:A2051, U:A2613 |
|
A:A2614, A:A2051 |
|
4dhc |
A3632 |
4Pout·2Bout |
|
|
|
A4510, A4511, A4512, A4513, A4514, A4515 |
|
G:A2578, A:A2614, A:A2051, U:A2613 |
|
A:A2614, A:A2051 |
|
4dr5 |
A1838 |
4Pout·2Bout |
|
|
|
A2414, A2415, A2416, A2417, A2418, A2419 |
|
U:A757, G:A758, U:A580, G:A581 |
|
G:A581, G:A758 |
|
4dr6 |
A1886 |
4Pout·2Bout |
|
|
|
A2778, A2779, A2780, A2781, A2782, A2783 |
|
U:A757, G:A758, U:A580, G:A581 |
|
G:A581, G:A758 |
|