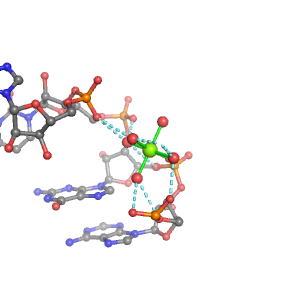
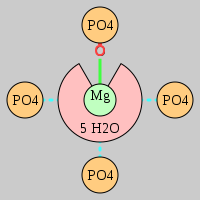
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4tpf |
A3074 |
Oph·3Pout |
A:A2518(OP2) |
|
|
A3533, A3534, A3535, A3536, A3537 |
|
U:A2519, C:A2540, A:A2541 |
C:A2517 |
A:A2541 |
|
4tpf |
A3044 |
Oph·3Pout |
G:A1333(OP2) |
|
|
A3388, A3389, A3390, A3391, A3392 |
|
U:A1313, G:A1334, G:A1332 |
|
C:A1335 |
|
4tpf |
A3070 |
Oph·3Pout |
A:A2270(OP2) |
|
|
A3512, A3513, A3514, A3515, A3516 |
|
G:A2269, A:A2267, G:A2271 |
|
A:A2270, G:A2271, U:A2272 |
|
4tpf |
A3131 |
Oph·3Pout |
A:A1009(OP2) |
|
|
A3777, A3778, A3779, A3780 |
|
A:A1010, C:A1006, C:A1007 |
|
A:A1010 |
|
4tpf |
A3122 |
Oph·3Pout |
G:A1828(OP2) |
|
|
A3742, A3743, A3744, A3745, C306 |
|
U:A1827, G:A1972, C:A1788 |
|
G:A1828 |
|
4tpf |
A3066 |
Oph·3Pout |
A:A2062(OP1) |
|
|
A3493, A3494, A3495, A3496, A3497 |
|
G:A2061, C:A2443, G:A2444 |
|
G:A2444, G:A2445 |
|
4tpf |
A3050 |
Oph·3Pout |
U:A1647(OP2) |
|
|
A3415, A3416, A3417, A3418, A3419 |
|
A:A1272, A:A1618, C:A1617 |
C:A1646, A:A1616 |
G:A1271 |
|
4tpf |
A3033 |
Oph·3Pout |
C:A946(OP2) |
|
|
A3344, A3345, A3346, A3347 |
|
A:A945, G:A822, C:A823 |
A:A821, C:A944 |
|
|
4tpf |
A3006 |
Oph·3Pout |
A:A218(OP2) |
|
|
A3223, A3224, A3225, A3226, A3227 |
|
A:A217, U:A427, A:A428 |
|
A:A218, A:A219, G:A220 |
|
4tpe |
A1619 |
Oph·3Pout |
C:A578(OP1) |
|
|
A1786, A1787, A1788, A1789, A1790 |
|
C:A817, C:A576, U:A820 |
G:A575 |
|
|
4tpd |
A3043 |
Oph·3Pout |
G:A1333(OP2) |
|
|
A3392, A3393, A3394, A3395, A3396 |
|
C:A1314, G:A1334, G:A1332 |
|
G:A1334, G:A1333 |
|
4tpd |
A3007 |
Oph·3Pout |
G:A297(OP2) |
|
|
A3228, A3229, A3230, A3231, A3232 |
|
U:A296, G:A298, G:A338 |
|
G:A298, G:A297 |
|
4tpd |
A3118 |
Oph·3Pout |
G:A862(OP2) |
|
|
A3730, A3731, A3732, A3733, A3734 |
|
A:A861, A:A910, A:A863 |
|
G:A862, A:A863 |
|
4tpd |
A3073 |
Oph·3Pout |
A:A2518(OP2) |
|
|
A3537, A3538, A3539, A3540, A3541 |
|
U:A2519, C:A2540, A:A2541 |
C:A2517 |
A:A2541 |
|
4tpd |
A3009 |
Oph·3Pout |
G:A450(OP2) |
|
|
A3238, A3239, A3240, A3241, E301 |
|
U:A451, G:A452, A:A449 |
|
|
|
4tpd |
A3056 |
Oph·3Pout |
G:A1828(OP1) |
|
|
A3454, A3455, A3456, A3457, A3458 |
|
A:A1773, G:A1973, A:A1772 |
|
|
|
4tpd |
A3040 |
Oph·3Pout |
A:A2005(OP1) |
|
|
A3384, A3385, A3386, A3387 |
|
A:A1268, A:A1269, C:A2006 |
|
A:A1269 |
|
4tpc |
A1609 |
Oph·3Pout |
C:A578(OP1) |
|
|
A1739, A1740, A1741, A1742, A1743 |
|
C:A576, C:A817, U:A820 |
G:A575 |
|
|
4tpb |
A3065 |
Oph·3Pout |
A:A2062(OP1) |
|
|
A3492, A3493, A3494, A3495, A3496 |
|
G:A2061, C:A2443, G:A2444 |
|
G:A2444 |
|
4tpb |
A3130 |
Oph·3Pout |
A:A1009(OP2) |
|
|
A3776, A3777, A3778, A3779 |
|
A:A1010, C:A1006, C:A1007 |
|
A:A1010 |
|
4tpb |
A3121 |
Oph·3Pout |
G:A1828(OP2) |
|
|
A3741, A3742, A3743, A3744, C306 |
|
U:A1827, G:A1972, C:A1788 |
|
G:A1828 |
|
4tpb |
A3049 |
Oph·3Pout |
U:A1647(OP2) |
|
|
A3415, A3416, A3417, A3418, A3419 |
|
A:A1618, A:A1272, C:A1617 |
A:A1616, C:A1646 |
G:A1271 |
|
4tpb |
A3118 |
Oph·3Pout |
A:A1938(OP2) |
|
|
A3726, A3727, A3728, A3729, A3730 |
|
A:A1937, C:A1957, C:A1958 |
G:A1770 |
|
|
4tpb |
A3006 |
Oph·3Pout |
A:A218(OP2) |
|
|
A3223, A3224, A3225, A3226, A3227 |
|
A:A217, U:A427, A:A428 |
|
A:A218, A:A219, G:A220 |
|
4tpb |
A3054 |
Oph·3Pout |
C:A1774(OP1) |
|
|
A3440, A3441, A3442, A3443, A3444 |
|
U:A1775, A:A1787, A:A1786 |
A:A1772 |
G:A1776, C:A1788, A:A1789, A:A1787 |
|