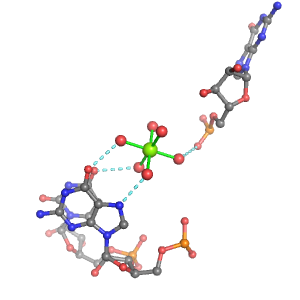
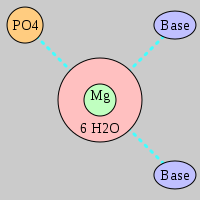
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
4gaq |
A1607 |
Pout·2Bout |
|
|
|
A1724, A1725, A1726, A1727, A1728, T101 |
|
G:A259 |
|
G:A260, G:A259 |
|
4gaq |
A1635 |
Pout·2Bout |
|
|
|
A1866, A1867, A1868, A1869, A1870, A1871 |
|
G:A1385 |
|
G:A1386, G:A1385 |
|
4gaq |
A1622 |
Pout·2Bout |
|
|
|
A1801, A1802, A1803, A1804, A1805, A1806 |
|
G:A803 |
|
G:A771, U:A772 |
|
4gar |
A3068 |
Pout·2Bout |
|
|
|
A3521, A3522, A3523, A3524, A3525 |
|
U:A2302 |
|
G:A2303, G:A2304 |
|
4gar |
A3113 |
Pout·2Bout |
|
|
|
A3716, A3717, A3718, T203 |
|
A:A1342 |
|
G:A1601, U:A1602 |
|
4gar |
A3082 |
Pout·2Bout |
|
|
|
A3582, A3583, A3584, A3585, A3586, A3587 |
|
A:A933 |
|
G:A841, U:A842 |
|
4gar |
A3015 |
Pout·2Bout |
|
|
|
A3278, A3279, A3280, A3281, A3282, A3283 |
|
G:A1256 |
|
U:A580, G:A1259 |
|
4gar |
A3131 |
Pout·2Bout |
|
|
|
A3794, A3795, A3796, A3797, A3798, 4201 |
|
A:A2741 |
|
C:A2755, G:A2742 |
|
4gau |
A3099 |
Pout·2Bout |
|
|
|
A3676, A3677, A3678, A3679, A3680, A3681 |
|
U:A2615 |
|
G:A2048, G:A2049 |
|
4gd1 |
A1602 |
Pout·2Bout |
|
|
|
A1706, A1707, A1708, A1709, A1710, A1711 |
|
A:A315 |
|
G:A318, G:A319 |
|
4gd2 |
A1606 |
Pout·2Bout |
|
|
|
A1724, A1725, A1726, A1727, A1728, A1729 |
|
G:A259 |
|
G:A259, G:A260 |
|
4ji0 |
A1815 |
Pout·2Bout |
|
|
|
A2131, A2132, A2133, A2134, A2135, A2136 |
|
C:A398 |
|
C:A36, U:A37 |
|
4ji0 |
A1639 |
Pout·2Bout |
|
|
|
A1971, A1972, A1973, A1974, A1975 |
|
A:A583 |
|
G:A584, G:A585 |
|
4ji1 |
A1731 |
Pout·2Bout |
|
|
|
A2076, A2077, A2078, A2079, A2080, A2081 |
|
A:A753 |
|
G:A650, G:A649 |
|
4ji3 |
A1698 |
Pout·2Bout |
|
|
|
A2065, A2066, A2067, N201 |
|
A:A1360 |
|
C:A980, A:A1360 |
|
4ji4 |
A1638 |
Pout·2Bout |
|
|
|
A1968, A1969, A1970, A1971, Q301 |
|
A:A583 |
|
G:A584, G:A585 |
|
4ji5 |
A1723 |
Pout·2Bout |
|
|
|
A2054, A2055, A2056, A2057, A2058, A2059 |
|
A:A781 |
|
G:A799, G:A800 |
|
4ji5 |
A1694 |
Pout·2Bout |
|
|
|
A1893, A1894, A1895, A1896, A1897, A1898 |
|
U:A871 |
|
G:A853, G:A854 |
|
4ji6 |
A1778 |
Pout·2Bout |
|
|
|
A2301, A2302, A2303, A2304, A2305, A2306 |
|
A:A753 |
|
G:A649, G:A650 |
|
4ji6 |
A1930 |
Pout·2Bout |
|
|
|
A3175, A3176, A3177, A3178, A3179, A3180 |
|
A:A781 |
|
G:A798, G:A800 |
|
4ji6 |
A1762 |
Pout·2Bout |
|
|
|
A2205, A2206, A2207, A2208, A2209, A2210 |
|
U:A871 |
|
G:A854, G:A853 |
|
4ji7 |
A1994 |
Pout·2Bout |
|
|
|
A3469, A3470, A3471, A3472, A3473, A3474 |
|
U:A646 |
|
C:A586, G:A587 |
|
4ji7 |
A1800 |
Pout·2Bout |
|
|
|
A2311, A2312, A2313, A2314, A2315, A2316 |
|
U:A871 |
|
G:A853, G:A854 |
|
4ji7 |
A1631 |
Pout·2Bout |
|
|
|
A2158, A2159, A2160, A2161, A2162 |
|
A:A583 |
|
G:A584, G:A585 |
|
4nvu |
A3165 |
Pout·2Bout |
|
|
|
A4048, A4049, A4050, A4051 |
|
G:A803 |
|
G:A771, U:A772 |
|