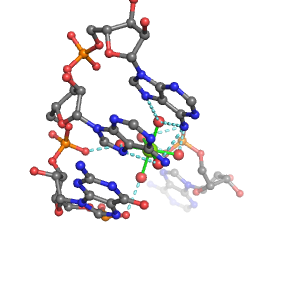
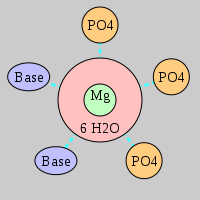
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
2qbg |
B2939 |
3Pout·2Bout |
|
|
|
B3188, B3189, B3190, B3191, B3192, B3193 |
|
A:B447, A:B28, U:B29 |
|
G:B473, G:B474 |
|
2qbg |
B2907 |
3Pout·2Bout |
|
|
|
B3027, B3028, B3029, B3030, B3031, B3032 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2qbg |
B2909 |
3Pout·2Bout |
|
|
|
B3037, B3038, B3039, B3040, B3041, B3042 |
|
A:B1936, G:B1959, A:B1937 |
|
A:B1960, G:B1959 |
|
2qbi |
B2953 |
3Pout·2Bout |
|
|
|
B3242, B3243, B3244, B3245, B3246, L284 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2qbi |
B3012 |
3Pout·2Bout |
|
|
|
B3495, B3496, B3497, B3498, B3499, B3500 |
|
G:B570, A:B973, U:B569 |
|
A:B820, A:B821 |
|
2qbj |
A1572 |
3Pout·2Bout |
|
|
|
A1755, A1756, A1757, A1758, A1759, A1760 |
|
A:A768, C:A805, A:A767 |
|
A:A768, G:A769 |
|
2qbj |
A1600 |
3Pout·2Bout |
|
|
|
A1883, A1884, A1885, A1886, A1887, A1888 |
|
A:A1394, G:A917, A:A918 |
|
A:A918, A:A919 |
|
2qbk |
B2907 |
3Pout·2Bout |
|
|
|
B3028, B3029, B3030, B3031, B3032, L145 |
|
U:B568, G:B831, C:B944 |
|
A:B945, A:B2448 |
|
2qbk |
B2909 |
3Pout·2Bout |
|
|
|
B3037, B3038, B3039, B3040, B3041, B3042 |
|
A:B1937, A:B1936, G:B1959 |
|
A:B1960, G:B1959 |
|
2qov |
B2931 |
3Pout·2Bout |
|
|
|
B3142, B3143, B3144, B3145, B3146, B3147 |
|
A:B1937, A:B1936, G:B1959 |
|
A:B1960, G:B1959 |
|
2qov |
B2953 |
3Pout·2Bout |
|
|
|
B3244, B3245, B3246, B3247, B3248, B3249 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2qov |
B2905 |
3Pout·2Bout |
|
|
|
B3015, B3016, B3017, B3018, B3019 |
|
A:B802, G:B674, A:B675 |
|
U:B803, A:B804 |
|
2qox |
B2907 |
3Pout·2Bout |
|
|
|
B3026, B3027, B3028, B3029, B3030, L145 |
|
U:B568, C:B944, G:B831 |
|
A:B945, A:B2448 |
|
2qoy |
A1550 |
3Pout·2Bout |
|
|
|
A1935, A1936, A1937, A1938, A1939, A1940 |
|
A:A315, A:A329, C:A316 |
|
G:A318, G:A319 |
|
2qp0 |
A1560 |
3Pout·2Bout |
|
|
|
A1975, A1976, A1977, A1978, A1979, A1980 |
|
A:A768, C:A805, A:A767 |
|
A:A768, G:A769 |
|
2qp1 |
B2987 |
3Pout·2Bout |
|
|
|
B3917, B3918, B3919, B3920, B3921, B3922 |
|
G:B2578, U:B2613, A:B2051 |
|
A:B2051, A:B2614 |
|
2z4l |
B2932 |
3Pout·2Bout |
|
|
|
B3141, B3142, B3143, B3144, B3145, B3146 |
|
A:B1937, A:B1936, G:B1959 |
|
A:B1960, G:B1959 |
|
2z4l |
B2954 |
3Pout·2Bout |
|
|
|
B3242, B3243, B3244, B3245, B3246, L284 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
2z4n |
B2908 |
3Pout·2Bout |
|
|
|
B3028, B3029, B3030, B3031, B3032, L145 |
|
U:B568, G:B831, C:B944 |
|
A:B945, A:B2448 |
|
2z4n |
B2910 |
3Pout·2Bout |
|
|
|
B3037, B3038, B3039, B3040, B3041, B3042 |
|
A:B1936, G:B1959, A:B1937 |
|
A:B1960, G:B1959 |
|
3df2 |
B3157 |
3Pout·2Bout |
|
|
|
B4158, B4159, B4160, B4161, B4162, B4163 |
|
A:B1937, A:B1936, G:B1959 |
|
A:B1960, G:B1959 |
|
3df2 |
B3282 |
3Pout·2Bout |
|
|
|
B4283, B4284, B4285, B4286, B4287, B4288 |
|
C:B944, G:B831, U:B568 |
|
A:B945, A:B2448 |
|
3df4 |
B3014 |
3Pout·2Bout |
|
|
|
B4015, B4016, B4017, B4019, B4020, L4018 |
|
U:B568, G:B831, C:B944 |
|
A:B945, A:B2448 |
|
3i1p |
A2968 |
3Pout·2Bout |
|
|
|
A3340, A3341, A3342, A3343, A3344, A3345 |
|
A:A2051, G:A2578, C:A2050 |
|
A:A2051, A:A2614 |
|
3i1t |
A2953 |
3Pout·2Bout |
|
|
|
A3267, A3268, A3269, A3270, A3271 |
|
G:A1456, A:A1453, C:A1454 |
|
G:A1456, U:A1457 |
|