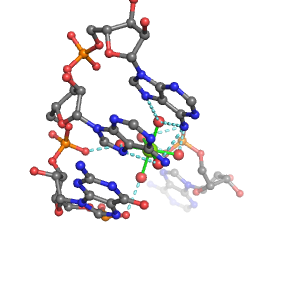
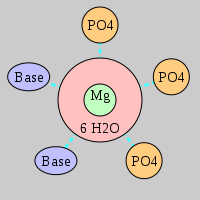
| Mg2+ ion |
Site type |
Inner-sphere ligands |
Outer-sphere ligands |
|---|
| PDB ID |
Mg2+ ID |
Oph |
Or |
Ob+Nb |
Water |
Other |
Pout |
Rout |
Bout |
|
3i20 |
A2916 |
3Pout·2Bout |
|
|
|
A3100, A3101, A3102, A3103, A3104, A3105 |
|
U:A568, G:A831, C:A944 |
|
A:A945, A:A2448 |
|
3i21 |
A2 |
3Pout·2Bout |
|
|
|
A1577, A1578, A1579, A1580, A1581, A1582 |
|
A:A315, A:A329, G:A319 |
|
A:A320, G:A319 |
|
3i22 |
A2962 |
3Pout·2Bout |
|
|
|
A3316, A3317, A3318, A3319, A3320, A3321 |
|
A:A1937, G:A1959, A:A1936 |
|
G:A1959, A:A1960 |
|
3ofq |
A2968 |
3Pout·2Bout |
|
|
|
A3342, A3343, A3344, A3345, A3346, A3347 |
|
A:A2051, G:A2578, C:A2050 |
|
A:A2051, A:A2614 |
|
3ofq |
A2986 |
3Pout·2Bout |
|
|
|
A3428, A3429, A3430, A3431 |
|
A:A217, A:A218, A:A428 |
|
A:A218, A:A219 |
|
3ofr |
A2917 |
3Pout·2Bout |
|
|
|
A3101, A3102, A3103, A3104, A3105, L145 |
|
C:A944, G:A831, U:A568 |
|
A:A945, A:A2448 |
|
3og0 |
A2966 |
3Pout·2Bout |
|
|
|
A3339, A3340, A3341, A3342, A3343, A3344 |
|
C:A2050, A:A2051, G:A2578 |
|
A:A2051, A:A2614 |
|
3orb |
A2994 |
3Pout·2Bout |
|
|
|
A3465, A3466, A3467, A3468, A3469, A3470 |
|
A:A1572, G:A1422, G:A1423 |
|
G:A1423, G:A1425 |
|
3owz |
A115 |
3Pout·2Bout |
|
|
|
A141, A142, A143, A144, A145, A146 |
|
U:A23, C:A24, G:A25 |
|
G:A25, G:A26 |
|
3r8s |
A2916 |
3Pout·2Bout |
|
|
|
A3158, A3159, A3160, A3161, A3162, L145 |
|
U:A568, G:A831, C:A944 |
|
A:A945, A:A2448 |
|
3r8t |
A2916 |
3Pout·2Bout |
|
|
|
A3128, A3129, A3130, A3131, A3132, L146 |
|
C:A944, G:A831, U:A568 |
|
A:A2448, A:A945 |
|
3v23 |
A3397 |
3Pout·2Bout |
|
|
|
A4259, A4260, A4261, A4262, A4263 |
|
G:A1332, G:A1334, U:A1313 |
|
G:A1334, U:A1335 |
|
3v23 |
B211 |
3Pout·2Bout |
|
|
|
B313, B314, B315, B316, B317 |
|
U:B74, A:B100, G:B75 |
|
G:B75, G:B76 |
|
3v23 |
A3599 |
3Pout·2Bout |
|
|
|
A4988, A4989, A4990, A4991, A4992, A4993 |
|
A:A1373, A:A1354, A:A1353 |
|
G:A1355, G:A1356 |
|
3v23 |
A3309 |
3Pout·2Bout |
|
|
|
A3920, A3921, A3922, A3923, A3924, P303 |
|
G:A831, G:A944, U:A568 |
|
A:A945, A:A2448 |
|
3v23 |
A3586 |
3Pout·2Bout |
|
|
|
A4916, A4917, A4918, A4919, A4920, A4921 |
|
A:A1632, G:A1633, G:A1630 |
|
C:A1631, G:A1630 |
|
3v23 |
A3345 |
3Pout·2Bout |
|
|
|
A4052, A4053, A4054, A4055, A4056, A4057 |
|
C:A1318, A:A1331, C:A1330 |
|
C:A1318, G:A1319 |
|
3v23 |
A3452 |
3Pout·2Bout |
|
|
|
A4458, A4459, A4460, A4461, A4462 |
|
U:A1357, C:A1370, G:A1358 |
|
G:A1358, G:A1371 |
|
3v25 |
A3264 |
3Pout·2Bout |
|
|
|
A3607, A3608, A3609, A3610, A3611 |
|
G:A1334, U:A1313, G:A1332 |
|
U:A1335, G:A1334 |
|
3v25 |
A3279 |
3Pout·2Bout |
|
|
|
A3670, A3671, A3672, A3673, A3674, P301 |
|
G:A831, G:A944, U:A568 |
|
A:A2448, A:A945 |
|
3v26 |
A1722 |
3Pout·2Bout |
|
|
|
A2000, A2001, A2002, A2003 |
|
A:A780, G:A799, G:A800 |
|
G:A800, A:A780 |
|
3v27 |
A3594 |
3Pout·2Bout |
|
|
|
A5046, A5047, A5048, A5049 |
|
G:A2532, A:A2533, A:A2534 |
|
A:A2534, G:A2535 |
|
3v27 |
A3324 |
3Pout·2Bout |
|
|
|
A3919, A3920, A3921, A3922, A3923, P301 |
|
G:A831, G:A944, U:A568 |
|
A:A945, A:A2448 |
|
3v27 |
A3451 |
3Pout·2Bout |
|
|
|
A4435, A4436, A4437, A4438, A4439, A4440 |
|
G:A1332, C:A1333, U:A1313 |
|
U:A1335, G:A1334 |
|
3v29 |
A3337 |
3Pout·2Bout |
|
|
|
A3768, A3769, A3770, A3771, A3772 |
|
U:A1313, G:A1334, G:A1332 |
|
U:A1335, G:A1334 |
|