X-ray Structures
Important note for CSGID staff! Before depositing your structure, make sure it follows the guidelines here " PDB deposit guidelines" and here "Crystal structure quality metrics for PDB deposition."
Remove FiltersThis search was filtered by:
Authors: Anderson
Authors: Anderson
Page 21 of 52, showing 20 records
out of 1038 total, starting on record 401 structures.
Click on the title of a structure below to see more information about that structure.
Target IDP90564 community request
Target IDP90922 community request
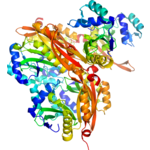
 Title: 1.85 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2.
Title: 1.85 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2.Authors:
Other ligands: 3,5-dihydro-5-methylidene-4h-imidazol-4-on
R: 17%
Rfree: 21.4%
Rfree: 21.4%
Resolution: 1.85Å
Deposited: 2009-12-15
Released: 2009-12-29
Released: 2009-12-29
Target IDP90258 community request
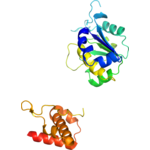 Title: 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252
Title: 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252Authors:
Other ligands: 3,5-dihydro-5-methylidene-4h-imidazol-4-on
R: 21.1%
Rfree: 25.5%
Rfree: 25.5%
Resolution: 2.35Å
Deposited: 2009-12-04
Released: 2009-12-22
Released: 2009-12-22
Target IDP90225 community request
Target IDP90163 community request
 Title: Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Reaction Intermediate.
Title: Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Reaction Intermediate. Authors:
Complexed with: 3,5-dihydro-5-methylidene-4h-imidazol-4-on
R: 19.2%
Rfree: 24.1%
Rfree: 24.1%
Resolution: 2.2Å
Deposited: 2009-09-09
Released: 2009-09-22
Released: 2009-09-22
Target IDP90224 community request
Target IDP00167 community request
 Title: Crystal structure of pre-cleavage form of cysteine protease domain from Vibrio cholerae RtxA toxin
Title: Crystal structure of pre-cleavage form of cysteine protease domain from Vibrio cholerae RtxA toxinAuthors:
Complexed with: inositol hexakisphosphate
Other ligands: chloride, Unknown atom
R: 17.1%
Rfree: 21.6%
Rfree: 21.6%
Resolution: 1.95Å
Deposited: 2009-01-26
Released: 2009-02-17
Released: 2009-02-17
Target IDP07236
 Title: 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid.
Title: 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid.Authors:
Complexed with:
Other ligands: , chloride, ethylene diol
R: 14.5%
Rfree: 15.5%
Rfree: 15.5%
Resolution: 1.3Å
Deposited: 2019-01-07
Released: 2019-01-16
Released: 2019-01-16
Target IDP07597
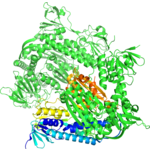 Title: 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
Title: 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.Authors:
Complexed with:
Other ligands: 2-[bis-(2-hydroxy-ethyl)-amino]-2-hydroxymethyl-propane-1,3-diol, bicarbonate ion, chloride, ethylene diol, riboflavine
R: 17.1%
Rfree: 21%
Rfree: 21%
Resolution: 1.9Å
Deposited: 2018-11-27
Released: 2018-12-12
Released: 2018-12-12
Target IDP07200
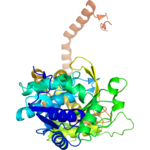 Title: 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
Title: 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.Authors:
Complexed with:
Other ligands: , chloride, ethylene diol, sulfate
R: 15.6%
Rfree: 17.9%
Rfree: 17.9%
Resolution: 1.5Å
Deposited: 2018-07-23
Released: 2018-08-01
Released: 2018-08-01
Target IDP01616
 Title: Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG
Title: Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeGAuthors:
Complexed with: n-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]aceta mide, spermine
Other ligands: 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, (4R)-2-methylpentane-2,4-diol, unknown, unknown
R: 12.5%
Rfree: 15.1%
Rfree: 15.1%
Resolution: 1.35Å
Deposited: 2018-07-10
Released: 2019-07-10
Released: 2019-07-10
Target IDP07306
 Title: 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD.
Title: 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD.Authors:
Complexed with: , formate
Other ligands: chloride
R: 17%
Rfree: 22.2%
Rfree: 22.2%
Resolution: 2.2Å
Deposited: 2018-06-01
Released: 2018-06-13
Released: 2018-06-13
Target IDP01047
 Title: 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
Title: 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.Authors:
Other ligands: chloride
R: 22.4%
Rfree: 26.4%
Rfree: 26.4%
Resolution: 2Å
Deposited: 2018-05-02
Released: 2018-05-16
Released: 2018-05-16
Target IDP01616
 Title: Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions.
Title: Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions.Authors:
Complexed with:
Other ligands: , , unknown
R: 17.3%
Rfree: 21.3%
Rfree: 21.3%
Resolution: 2.41Å
Deposited: 2018-04-02
Released: 2018-04-18
Released: 2018-04-18
Target IDP07215
 Title: 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine- diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium
Title: 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine- diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and MagnesiumAuthors:
Complexed with: , magnesium, uridine-diphosphate-2(n-acetylglucosaminyl) butyric aci d
Other ligands: chloride
R: 21.2%
Rfree: 25.2%
Rfree: 25.2%
Resolution: 2.75Å
Deposited: 2018-03-06
Released: 2018-03-21
Released: 2018-03-21